Sumeet Gupta
|
Sumeet Gupta |
Contents
Frequently Asked Questions
Solexa Data Processing
Is the Phix control useful for the analysis?
Yes. The control does allow us to distinguish between a cluster generation/sequencing issue vs a library/sample prep issue. In addition, it is useful to calculate the phasing and prephasing parameters (used to access the sequencing "efficiency") for the solexa base calling step which assumes a random distribution of bases. If we do not have a random distribution, it would lead to wrong base calls and worse quality scores, eventually leading to loss of data.
Do we filter "bad" reads from the final dataset? [06/19/09]
The default Illumina pipeline quality filter is used, which uses a threshold of CHASTITY >= 0.6. Chastity for a given base call is defined as "the ratio of the highest of the four (base type) intensities to the sum of highest two." This filter is used to identify clusters with a low signal to noise ratio, often as a result of two adjacent clusters being so physically close together that their signals cannot be measured independently.
We do NOT remove the filtered reads from the final dataset but we do mark each read ID with a boolean system indicating 1 for good/Not Filered reads and 0 for bad/filtered reads.
Was solexa pipeline version 1.3+ used for processing my data?
Use the "grep" command in unix to do a search for the characters unique to the Solexa older pipeline i.e. any of these characters in red below. If they are, then the data was generated using an older pipeline than 1.3.
...............................IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII......................
..........................XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
!"#$%&'()*+,-./0123456789:;<=>?@ABCDEFGHIJKLMNOPQRSTUVWXYZ[\]^_`abcdefghijklmnopqrstuvwxyz{|}~
| | | | | |
33 59 64 73 104 126
I - Illumina 1.3 Phred+64, 41 values (0, 40)
X - Solexa Solexa+64, 68 values (-5, 62)
Courtesy: http://en.wikipedia.org/wiki/FASTQ_format
Solexa Quality Scores
What is the format of the quality scores files (fasta files)?
@4:1:518:715
GATACCATAAAAGCTGGATCCTTCTTCAAGCATAA
+4:1:518:715
hhhhhhhhhhhhhhhdhhhhhhhhhhhdRehdhhP
@ID
Sequence
+ID
Quality Scores for each base (String of characters)
What do the quality scores for each base mean?
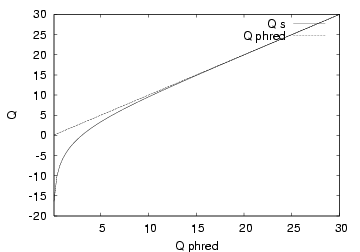
Old Solexa/Illumina pipeline FASTQ files used Solexa scores with an ASCII offset 64. New Solexa/Illumina 1.3+ pipeline FASTQ files use PHRED scores with an ASCII offset 64. These differ at the low end of the range of possible quality score with solexa scores allowing negative numbers. At Q scores above ~11 the two are very similar as indicated by the line plot below (Courtesy: Illumina).
!"#$%&'()*+,-./0123456789:;<=>?@ABCDEFGHIJKLMNOPQRSTUVWXYZ[\]^_`abcdefghijklmnopqrstuvwxyz{|}~
| | | | | |
33 59 64 73 104 126
Illumina 1.3 Phred+64, 41 values (0, 40)
Solexa Solexa+64, 68 values (-5, 62)
Courtesy: http://en.wikipedia.org/wiki/FASTQ_format
P(error) - Probability of the base call being incorrect.
| Char | ASCII | Char-64 | P(error) |
| ; | 59 | -5 | 0.7597 |
| < | 60 | -4 | 0.7153 |
| = | 61 | -3 | 0.6661 |
| > | 62 | -2 | 0.6131 |
| ? | 63 | -1 | 0.5573 |
| @ | 64 | 0 | 0.5 |
| A | 65 | 1 | 0.4427 |
| B | 66 | 2 | 0.3869 |
| C | 67 | 3 | 0.3339 |
| D | 68 | 4 | 0.2847 |
| E | 69 | 5 | 0.2403 |
| F | 70 | 6 | 0.2008 |
| G | 71 | 7 | 0.1663 |
| H | 72 | 8 | 0.1368 |
| I | 73 | 9 | 0.1118 |
| J | 74 | 10 | 0.0909 |
| K | 75 | 11 | 0.0736 |
| L | 76 | 12 | 0.0594 |
| M | 77 | 13 | 0.0477 |
| N | 78 | 14 | 0.0383 |
| O | 79 | 15 | 0.0307 |
| P | 80 | 16 | 0.0245 |
| Q | 81 | 17 | 0.0196 |
| R | 82 | 18 | 0.0156 |
| S | 83 | 19 | 0.0124 |
| T | 84 | 20 | 0.0099 |
| U | 85 | 21 | 0.0079 |
| V | 86 | 22 | 0.0063 |
| W | 87 | 23 | 0.005 |
| X | 88 | 24 | 0.004 |
| Y | 89 | 25 | 0.0032 |
| Z | 90 | 26 | 0.0025 |
| [ | 91 | 27 | 0.002 |
| \ | 92 | 28 | 0.0016 |
| ] | 93 | 29 | 0.0013 |
| ^ | 94 | 30 | 0.001 |
| _ | 95 | 31 | 0.0008 |
| ` | 96 | 32 | 0.0006 |
| a | 97 | 33 | 0.0005 |
| b | 98 | 34 | 0.0004 |
| c | 99 | 35 | 0.0003 |
| d | 100 | 36 | 0.0003 |
| e | 101 | 37 | 0.0002 |
| f | 102 | 38 | 0.0002 |
| g | 103 | 39 | 0.0001 |
| h | 104 | 40 | 0.0001 |
Is solexa quality same as phred quality scores?
The Solexa quality and Phred quality are asymptotically identical but NOT the same. If the error probability of a base is $e, the Solexa quality $sQ is: $sQ = -10 * log($e / (1 - $e)) / log(10);
Solexa quality $sQ can be converted to Phred quality $Q with this formula: $Q = 10 * log(1 + 10 ** ($sQ / 10.0)) / log(10);
Alignment of Next Generation Sequencing Reads
What is Eland?
ELAND stands for E fficient L arge-Scale A lignment of N ucleotide D atabases. ELAND is a alignment tool developed by Illumina/Solexa which searches a set of large DNA files for a large number of short DNA reads allowing up to 2 errors per match.
How can Eland be faster relative to some other alignment programs?
Given a sequence of length N, it can be divided into four subsequences (A, B, C, and D), which are of equal (or nearly equal length). Assuming there are no more than 2 errors, at least two of these subsequences will be "error free", so that the two error free sequences can then be searched for in a database containing all possible subsequences in the genome of interest. Thus, you can search your database for the subsequence AB and CD. Searching for the {AB and CD} subsequences would only work if the first half of your sequence has no errors. What if B and C had the errors? The answer is to shuffle your subsequences to make other combinations: ({AB and CD}, {AC and BD}, {AD and CD}, {BA and CD}, etc.). This still provides you with a relatively small search space for each sequence, as there are only 4! possible combinations (which is 4x3x2x1 = 24 possible sequences) to search for. This can be bound even further because the first pair and second pair are in the correct order, (ie {AB and CD} and {AC and BD} are ok, but {CA and DB} and {BA and DC} would give you an incorrect result) limiting you to only six possible combinations, which still allows you to find any correct match where at least two of the four subsequences are error free.
Combining these subsequences into 2 subsets rather than searching for 4 independent entries in their database speeds up the process as long as one set matches (matching criteria on the other set can be relaxed, ie, allowing for mismatches). That is to say, if you make two sequences out of the 4 subsequences {AB and CD}, you can search for an exact match for AB, and test all the possible results for mismatches in the {CD} portion of the sequence. This has the effect of significantly reducing the search space. (Only sequences containing the exact match to {AB})
Are their other alignment programs?
- Iterative ELAND - This algorithm, developed at the core by Sumeet Gupta, iteratively checks shorter reads to attempt to recover non-matching reads. (Currently requests are made to Sumeet Gupta for alignments but a stable version would soon be released which would also report positions if read maps at multiple positions).
- MAQ - Maq stands for Mapping and Assembly with Quality. It builds assembly by mapping short reads to reference sequences. Maq is a project hosted by SourceForge.net. The project page is available at http://sourceforge.net/projects/maq/.
- Bowtie - Bowtie is an ultrafast, memory-efficient short read aligner. It indexes the genome with a Burrows-Wheeler index to keep its memory footprint small: typically about 2.2 GB for the human genome (2.9 GB for paired-end). It supports alignment policies equivalent to Maq and SOAP but is substantially faster.
File Formats
- QC File Format Based on Quality Scores - http://jura.wi.mit.edu/genomecorewiki/index.php/QCOutputFormat