Difference between revisions of "SequencingQC"
From Genome Technology Core (GTC) wiki - Sequencing and Microarray
| Line 1: | Line 1: | ||
Note: We do not remove the reads that are suppose to be filtered by the solexa pipeline in version 1.4. However, reads suppose to be filtered are marked using a binary system of 1 = Good/Not Filtered and 0 = Bad/Filtered within the read ID. This information is in the quality score/FASTQ files. | Note: We do not remove the reads that are suppose to be filtered by the solexa pipeline in version 1.4. However, reads suppose to be filtered are marked using a binary system of 1 = Good/Not Filtered and 0 = Bad/Filtered within the read ID. This information is in the quality score/FASTQ files. | ||
| − | == '''''Sequencing Quality Control Based On FASTQ (Basecalls and quality scores)''''' | + | == '''''Sequencing Quality Control Based On FASTQ (Basecalls and quality scores)''''' || '''''[[ELANDQC|Go to Sequencing Quality Control Based On ELAND Alignments]]''''' == |
| − | |||
| − | |||
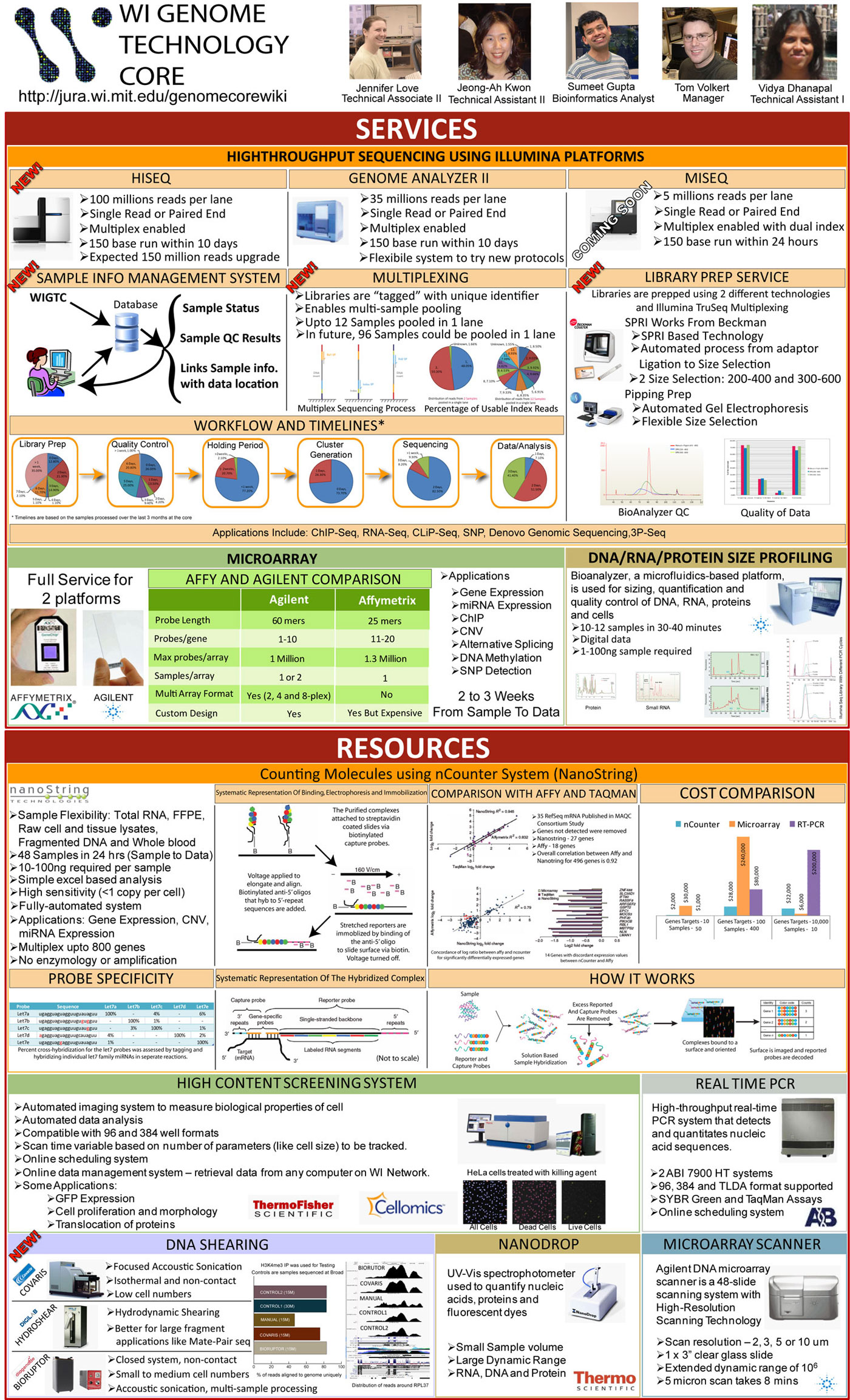
[[Image:WIGTC-2011-retreat-poster.jpg]] | [[Image:WIGTC-2011-retreat-poster.jpg]] | ||
Revision as of 12:13, 10 November 2011
Note: We do not remove the reads that are suppose to be filtered by the solexa pipeline in version 1.4. However, reads suppose to be filtered are marked using a binary system of 1 = Good/Not Filtered and 0 = Bad/Filtered within the read ID. This information is in the quality score/FASTQ files.